Mob/ Whatsapp/Wechat: +86 138 806 76215 | Email: [email protected] / [email protected]
 The DNA identification system 30A uses six-color fluorescent labeling technology to amplify 29 autosomal S TR loci , one Amel locus and one Yindel locus at one time
The DNA identification system 30A uses six-color fluorescent labeling technology to amplify 29 autosomal S TR loci , one Amel locus and one Yindel locus at one time suitable for amplification after sample extraction, and can also directly amplify blood stain or saliva spot samples on FTA cards,which is no need to use extraction kit .
suitable for amplification after sample extraction, and can also directly amplify blood stain or saliva spot samples on FTA cards,which is no need to use extraction kit .The DNA identification system 30A uses six-color fluorescent labeling technology to amplify 29 autosomal S TR loci , one Amel locus and one Yindel locus at one time, and can directly amplify filter paper, F TA card, cotton swab , saliva card, oral swab.
1. After receiving the kits frozen in dry ice or gel ice packs, if they are not used temporarily, please store them below -20°C;
2. After the kit is taken out for use, the pre-reaction component 4× premix C2 is stored at -20°C, and the remaining pre-reaction reagents are stored at 4°C to avoid repeated freezing and thawing. If the single dosage is small, it is recommended to store at -20°C after aliquoting ;
3. Store the components after the reaction at 4°C, avoid repeated freezing and thawing, and do not touch the reagents before the reaction to avoid contamination.
1. amount of DNA template
The concentration of DNA typing standard 9948 in this kit is 2ng/ μl . Users can properly adjust the number of cycles in the PCR program according to the amount of their DNA template. If the amount of template is too low, some alleles may be missing, and if the amount of template is too high, it may lead to results beyond the detection range of the genetic analyzer.
2. Shaking and mixing of reagents
the 4× master mix C2 and 5×30AM primer mixture for 10 seconds before use, and then centrifuge at low speed to rule out the uneven concentration of the solution caused by possible tube wall adsorption.
3. Amplification system preparation
Table1: Standard (extraction-free) amplification reaction system preparation composition
components | 25 μl system ( μl ) | 10 μl system ( μl ) | |
Master Mix | 4× Premix C2 | 6.25 | 2.5 |
5 × 30A M Primer Mix | 5.0 | 2 | |
Deionized water | 13.75 | 5.5 | |
bloodstain | Diameter 1.2mm | Diameter 1.2mm | |
total reaction volume | 25 μl | 10 μl | |
4. Protocol settings
Table2: PCR amplification procedure
transsexual | 95°C, 2 minutes |
28± 1 cycle | 94°C, 5 seconds |
60°C, 2 minutes | |
extend | 60°C, 2 minutes |
Cryopreservation | 15°C |
Note: If using ABI 9700 (Gold Block) or Veriti thermal cycler, please perform amplification in MAX mode.
1. Make sure that the POP4 gel and Buffer of the sequencer are within the validity period ;
2. Mix the 6 -color Matrix spectral calibration reagent and deionized formamide according to the following ratio , vortex and mix, and distribute to 96-well plate , 10 μl per well;
Instrument type | 3130 x l | 3500 | 3 500 xl |
6-color Matrix | 8 μl | 2.7 μl | 5 μl |
Deionized formamide | 200μl | 100 μl | 250μl _ |
3. After denaturation at 95°C for 3 minutes , quickly cool at 0°C for 3 minutes;
4. Spectral Calibration Recommended Parameters
4.1. 3130 series: ①Create a Run Module: Name it GoldeneyeJ6Matrix, select SPECTRAL for Type, and Spect36_POP4 for Template ; in Run Module Settings: fill in 3 for Injection_Voltage , fill in 10 for Injection_Time , fill in 1500 for Run_Time , and fill in other parameters according to default values. ②Create Protocol: Name as Golden Eye yeJ6, select J6 for Dye Set, and select Golden Eye yeJ6Matrix for Run Module ; in Edit Parameter, fill in 2.0 for Lower of Matrix Condition Number Bounds, and fill in 20 for Upper.
4.2. 3500 series: Create Dye Set : Dye Set Name is named GoldenEye J6, Chemistry selects Matrix Standard, Dye Set Template selects J6 Template ; in the Parameters option, Matrix Condition Number Upper Limit is set to 8.0 (adjustable according to actual conditions), and After Scan is set Subtract 200 from the Scan point value of the first fragment peak of the matrix , add 200 to the Scan point value of the largest fragment peak of the matrix in Before Scan, and fill in other parameters with default values.
5. In order to obtain the best correction effect, it is recommended to set the Sensitivity to 0.4, the Minimum Quality Score to 0.95 , and at the same time ensure that the spectral correction peak height is between 750rfu-4000rfu (3 130 series) or 3000rfu-20000rfu ( 3 500 series) . If this standard cannot be reached, it can be adjusted according to the actual electrophoresis situation.
1. Configure the sample loading system according to the ratio of molecular weight internal standard : deionized formamide = 0.5μl : 9.5μl , and distribute 10μl/ well ;
2. Amount of PCR product used: 1 μl , Amount of Allelic Ladder used: 1 μl
3. After denaturation at 95°C for 3 minutes , quickly cool at 0°C for 3 minutes;
4. Electrophoresis detection: The recommended injection voltage of 3130 series is 3k Volts, and the injection time is 10 sec; the injection voltage of 3500 series is 1.2k Volts, and the injection time is 24 sec.
1. Import panels and bin files , establish Analysis Method and Size Standard (JR 500: 65, 70, 80, 100, 120, 140, 160, 180, 200, 225, 250, 275, 300, 330, 360, 390, 420, 450, 490, 500);
2. Import the electrophoresis data, and select the corresponding analysis parameters such as panel, analysis method and size standard to analyze the data.
Reagent test kit | component name | Volume ( μl /tube) | Quantity (tube) |
Pre-reaction Kit | 4× Premix C2 | 625 | 2 |
5 x 30 A M primer mix | 500 | 2 | |
DNA Typing Standards 9948 (2ng/ μl ) | 2 5 | 1 | |
Deionized water | 1700 | 2 | |
Post-reaction kit | 30A Ladder _ | 40 | 1 |
Molecular weight internal standard JR 500 | 150 | 2 |
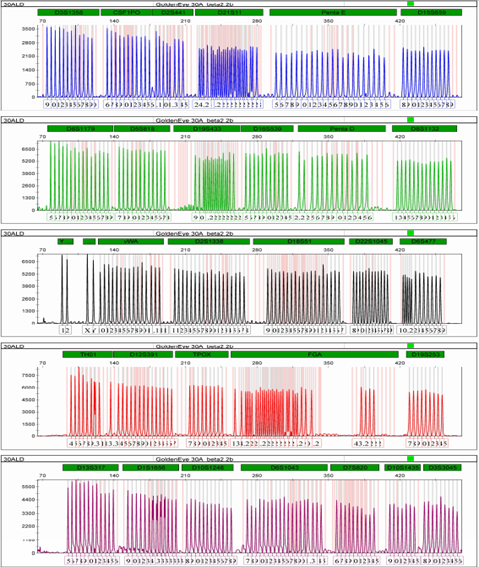
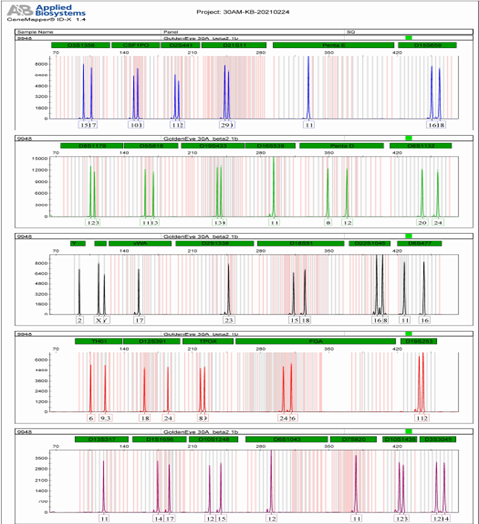
attached 2: Kit locus information
loci | Group | Control DNA | allele |
D3S1358 | B | 15,17 | 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, |
CSF1PO | B | 10,11 | 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, |
D2S441 | B | 11,12 | 8.1, 9, 10, 11, 11.3, 13, 14, 15, |
D21S11 | B | 29,30 | 24, 24.2, 25, 26, 27, 27.2, 28, 28.2, 29, 29.2, 30, 30.2, 31, 31.2, 32, 32.2, 33, 33.2, 34, 34.2, 35, 35.2, 36.2, 37, 38,, |
Penta E | B | 11 | 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26 |
D15S659 | B | 16,18 | 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, |
D8S1179 | G | 12,13 | 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, |
D5S818 | G | 11,13 | 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, |
D19S433 | G | 13,14 | 9, 10, 11, 11.2, 12,12.2, 13, 13.2, 14, 14.2, 15, 15.2, 16, 16.2, 17, 17.2, 18.2, |
D16S539 | G | 11 | 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, |
Penta D | G | 8,12 | 2.2, 3.2, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, |
D8S1132 _ | G. | 2 0.24 | 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24,25, 26, |
yindel | AND | 2 | 1, 2, |
Amel | AND | X,Y | X,Y, |
vWA | AND | 17 | 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22.1, 23.1, 24.1, |
D2S1338 | AND | 23 | 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, |
D18S51 | AND | 15.18 | 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, |
D22S1045 | Y | 16,18 | 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, |
D6S477 | Y | 11,16 | 10, 10.2, 11, 11.2, 12, 13, 14, 15, 16, 17, 18, 19, |
TH01 | R | 6,9.3 | 4, 5, 6, 7, 8, 9, 9.3, 10, 11, 13.3, |
D12S391 | R | 18,24 | 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, |
TPOX | R | 8,9 | 7, 8, 9, 10, 11, 12, 13, 14, 15, |
FGA | R | 24,26 | 13, 14.2, 15.2, 16, 16.2, 17, 18, 18.2, 19, 19.2, 20, 20.2, 21, 21.2, 22, 22.2, 23, 23.2, 24, 24.2, 25.2, 26, 27, 27.2, 28, 29, 30, 31.2, 43.2, 44.2, 45.2, 46.2, |
D19S253 | R | 11,12 | 7, 8, 9, 10, 11, 12, 13, 14, 15, |
D13S317 | P | 11 | 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, |
D1S1656 | P | 14,17 | 9, 10, 11, 12, 13, 14, 14.3, 15, 15.3, 16, 16.3, 17, 17.3, 18, 18.3, 19.3, 20.3, |
D10S1248 | P | 12,15 | 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, |
D6S1043 | P | 12 | 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 21.3, 23, 24, 25, |
D7S820 | P | 11 | 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, |
D 10S1435 | P | 1 2,13 | 9, 10, 11, 12, 13, 14, 15, |
D 3S3045 | P | 1 2,14 | 8, 9, 10, 11, 12, 13, 14, 15, 16, |
Notice:
1. This kit is valid for one year, see the product packaging for the production date and batch number
2. Can be used for family identification, forensic research, scientific research, etc.
3. The trial pack is for 25 people, the attached table is the composition list for 200 people, and the trial pack comes with spectral correction!